Predict Patient Risk Based on Gene Expression Data
Source:R/predictPatientRisk.R
predict_PatientRisk.RdFunction to predict the risk for new patients considering the gene expression for a subset of genes and the multivariate Cox regression model trained by function patientRisk, it may be used to predict over a single patient.
predict_PatientRisk(model.fit, mExpr.testData)Arguments
- model.fit
A list containing the pre-fitted model and necessary parameters for risk prediction, including the optimal lambda value, risk thresholds, and plot values.
- mExpr.testData
A data frame representing the gene expression data of test patients, where each row is a gene and each column is a sample.
Value
A list containing the following elements:
- risk_score: A vector with the unscaled risk score for new patients
estimated by a multivariate Cox regression model.
- scaled_risk_score: A vector with the risk score for the new
patients scaled to be interpretable in the range 0-100.
- plot_values: A list containing information for visualizing the
sigmoid curve and risk thresholds.
Details
A risk score is estimated for new patients considering the optimal regularized multivariate Cox regression model trained by the function patientRisk. The risk score is normalized to be interpretable in the scale (0-100). The function generates a risk plot for new patients and stratifies them in three risk groups (low, intermediate, high) considering the thresholds learned by function patientRisk.
References
martinezromero2018asuri BuenoFortes2023asuri
Examples
data(seBRCA)
# prefilterSAM ---
groupsVector <- SummarizedExperiment::colData(seBRCA)$ER.IHC
set.seed(5)
DE_list_genes <- prefilterSAM(seBRCA, groupsVector)
#> 2025-07-08 12:24:03.228718
#>
|
| | 0%
|
| | 1%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|= | 2%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|== | 3%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|== | 4%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|== | 5%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=== | 6%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==== | 7%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==== | 8%
|
|==== | 9%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|===== | 10%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====== | 11%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====== | 12%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====== | 13%
|
|======= | 14%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======== | 15%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======== | 16%
|
|======== | 17%
|
|========= | 18%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========== | 19%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========== | 20%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========== | 21%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=========== | 22%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============ | 23%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============ | 24%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============ | 25%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============= | 26%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============== | 27%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============== | 28%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============== | 29%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=============== | 30%
|
|================ | 31%
|
|================ | 32%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================ | 33%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================= | 34%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================== | 35%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================== | 36%
|
|================== | 37%
|
|=================== | 38%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================== | 39%
|
|==================== | 40%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================== | 41%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|===================== | 42%
|
|====================== | 43%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====================== | 44%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====================== | 45%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================= | 46%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================== | 47%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================== | 48%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================== | 49%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========================= | 50%
|
|========================== | 51%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========================== | 52%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========================== | 53%
|
|=========================== | 54%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================ | 55%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================ | 56%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================ | 57%
|
|============================= | 58%
|
|============================== | 59%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================== | 60%
|
|============================== | 61%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=============================== | 62%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================ | 63%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================ | 64%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================ | 65%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================= | 66%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================== | 67%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================== | 68%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================== | 69%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=================================== | 70%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================================== | 71%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================================== | 72%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================================== | 73%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|===================================== | 74%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====================================== | 75%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====================================== | 76%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|====================================== | 77%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================================= | 78%
|
|======================================== | 79%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|======================================== | 80%
|
|======================================== | 81%
|
|========================================= | 82%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========================================== | 83%
|
|========================================== | 84%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|========================================== | 85%
|
|=========================================== | 86%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================ | 87%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================ | 88%
|
|============================================ | 89%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================= | 90%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================== | 91%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================== | 92%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|============================================== | 93%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|=============================================== | 94%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================================ | 95%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================================ | 96%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================================ | 97%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|================================================= | 98%
#> Warning: The spline based estimation of pi0 results in a non-positive value of pi0.
#> Therefore, pi0 is estimated by using lambda = 0.5.
#>
|
|==================================================| 99%
|
|==================================================| 100%
#> 2025-07-08 12:25:30.658604
# genePheno ---
vectorSampleID <- rownames(SummarizedExperiment::colData(seBRCA))
vectorGroups <- SummarizedExperiment::colData(seBRCA)$ER.IHC |> as.numeric()
Pred_ER.IHC <- genePheno(seBRCA, DE_list_genes, vectorGroups, vectorSampleID)
#>
|
| | 0%
|
| | 1%
|
|= | 2%
|
|== | 3%
|
|== | 4%
|
|== | 5%
|
|=== | 6%
|
|==== | 7%
|
|==== | 8%
|
|==== | 9%
|
|===== | 10%
|
|====== | 11%
|
|====== | 12%
|
|====== | 13%
|
|======= | 14%
|
|======== | 15%
|
|======== | 16%
|
|======== | 17%
|
|========= | 18%
|
|========== | 19%
|
|========== | 20%
|
|========== | 21%
|
|=========== | 22%
|
|============ | 23%
|
|============ | 24%
|
|============ | 25%
|
|============= | 26%
|
|============== | 27%
|
|============== | 28%
|
|============== | 29%
|
|=============== | 30%
|
|================ | 31%
|
|================ | 32%
|
|================ | 33%
|
|================= | 34%
|
|================== | 35%
|
|================== | 36%
|
|================== | 37%
|
|=================== | 38%
|
|==================== | 39%
|
|==================== | 40%
|
|==================== | 41%
|
|===================== | 42%
|
|====================== | 43%
|
|====================== | 44%
|
|====================== | 45%
|
|======================= | 46%
|
|======================== | 47%
|
|======================== | 48%
|
|======================== | 49%
|
|========================= | 50%
|
|========================== | 51%
|
|========================== | 52%
|
|========================== | 53%
|
|=========================== | 54%
|
|============================ | 55%
|
|============================ | 56%
|
|============================ | 57%
|
|============================= | 58%
|
|============================== | 59%
|
|============================== | 60%
|
|============================== | 61%
|
|=============================== | 62%
|
|================================ | 63%
|
|================================ | 64%
|
|================================ | 65%
|
|================================= | 66%
|
|================================== | 67%
|
|================================== | 68%
|
|================================== | 69%
|
|=================================== | 70%
|
|==================================== | 71%
|
|==================================== | 72%
|
|==================================== | 73%
|
|===================================== | 74%
|
|====================================== | 75%
|
|====================================== | 76%
|
|====================================== | 77%
|
|======================================= | 78%
|
|======================================== | 79%
|
|======================================== | 80%
|
|======================================== | 81%
|
|========================================= | 82%
|
|========================================== | 83%
|
|========================================== | 84%
|
|========================================== | 85%
|
|=========================================== | 86%
|
|============================================ | 87%
|
|============================================ | 88%
|
|============================================ | 89%
|
|============================================= | 90%
|
|============================================== | 91%
|
|============================================== | 92%
|
|============================================== | 93%
|
|=============================================== | 94%
|
|================================================ | 95%
|
|================================================ | 96%
|
|================================================ | 97%
|
|================================================= | 98%
|
|==================================================| 99%
|
|==================================================| 100%
# Survival times should be provided in YEARS
time <- 'time'
status <- 'status'
# Pred_ER.IHC$genes is the subset of genes to be tested. In our case study,
# it is the list of genes related to the ER clinical variable that was
# obtained using the function **genePheno()**.
geneList <- names(Pred_ER.IHC$genes)
# Training of the multivariate COX model. Provide the expression matrix
# (genes as rows and samples as columns) for the list of genes selected,
# the time and the status vectors, and the method to stratify the patients
# (select one of these methods: `min.pval`, `med.pval`, `class.probs`).
set.seed(5)
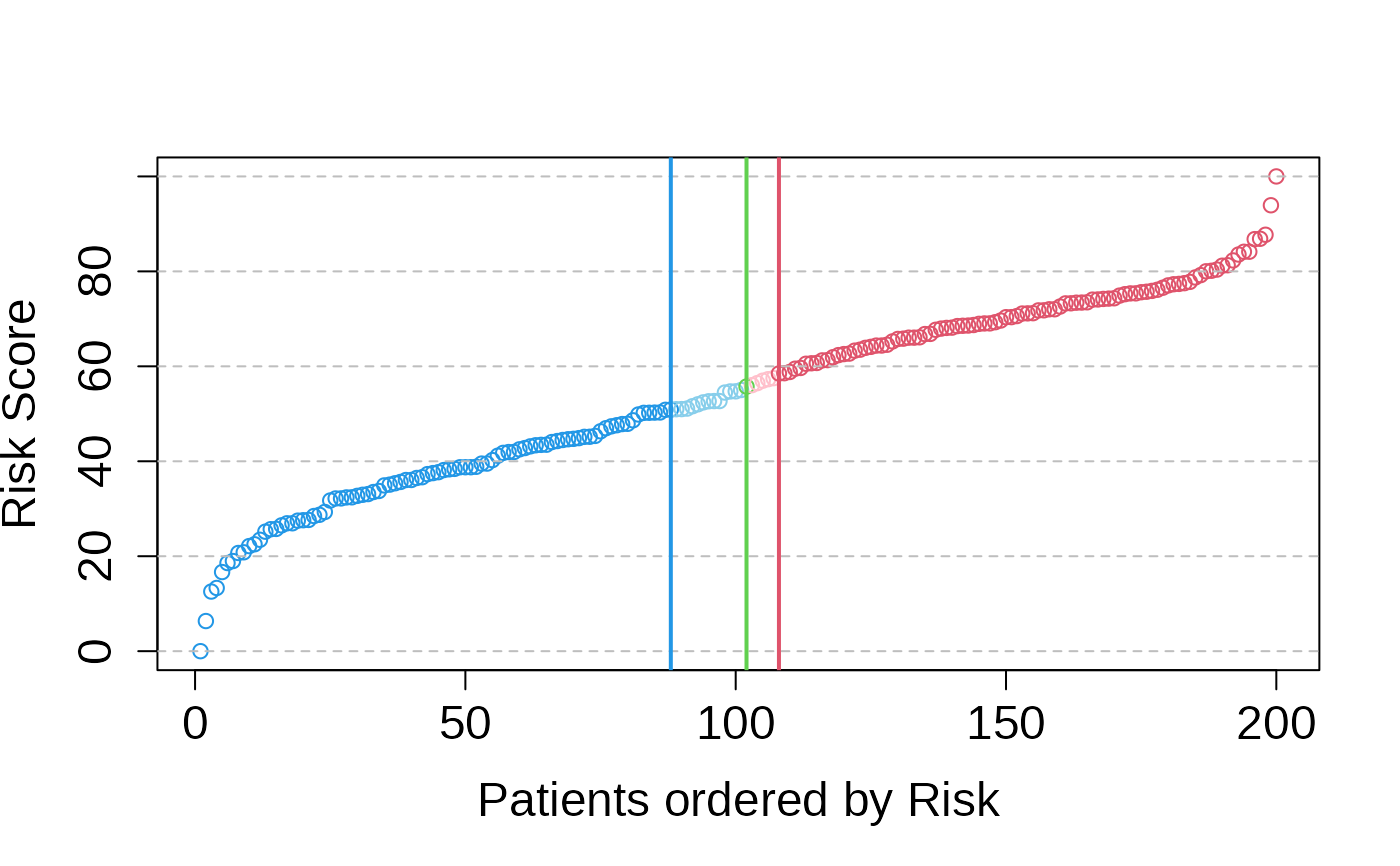
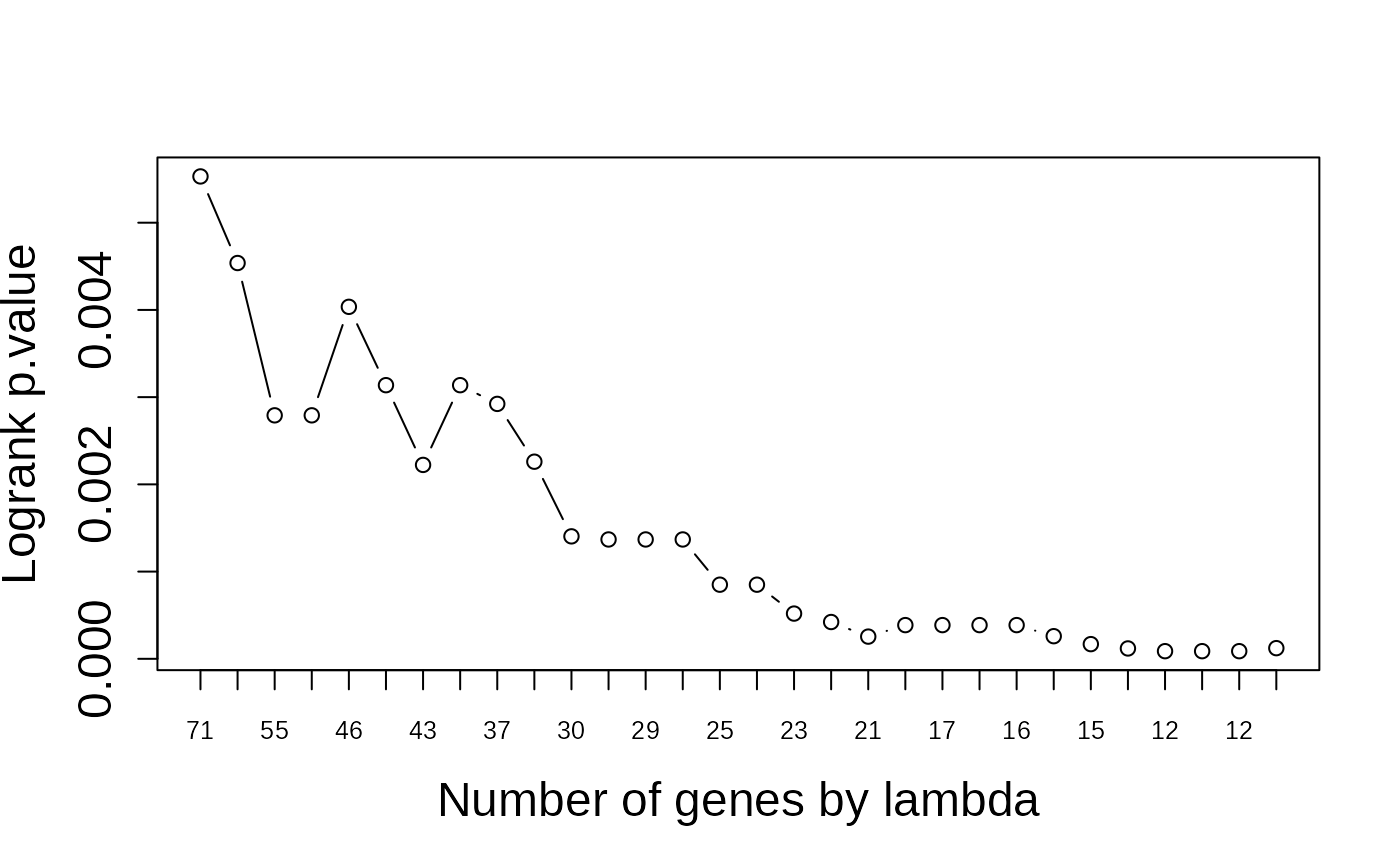
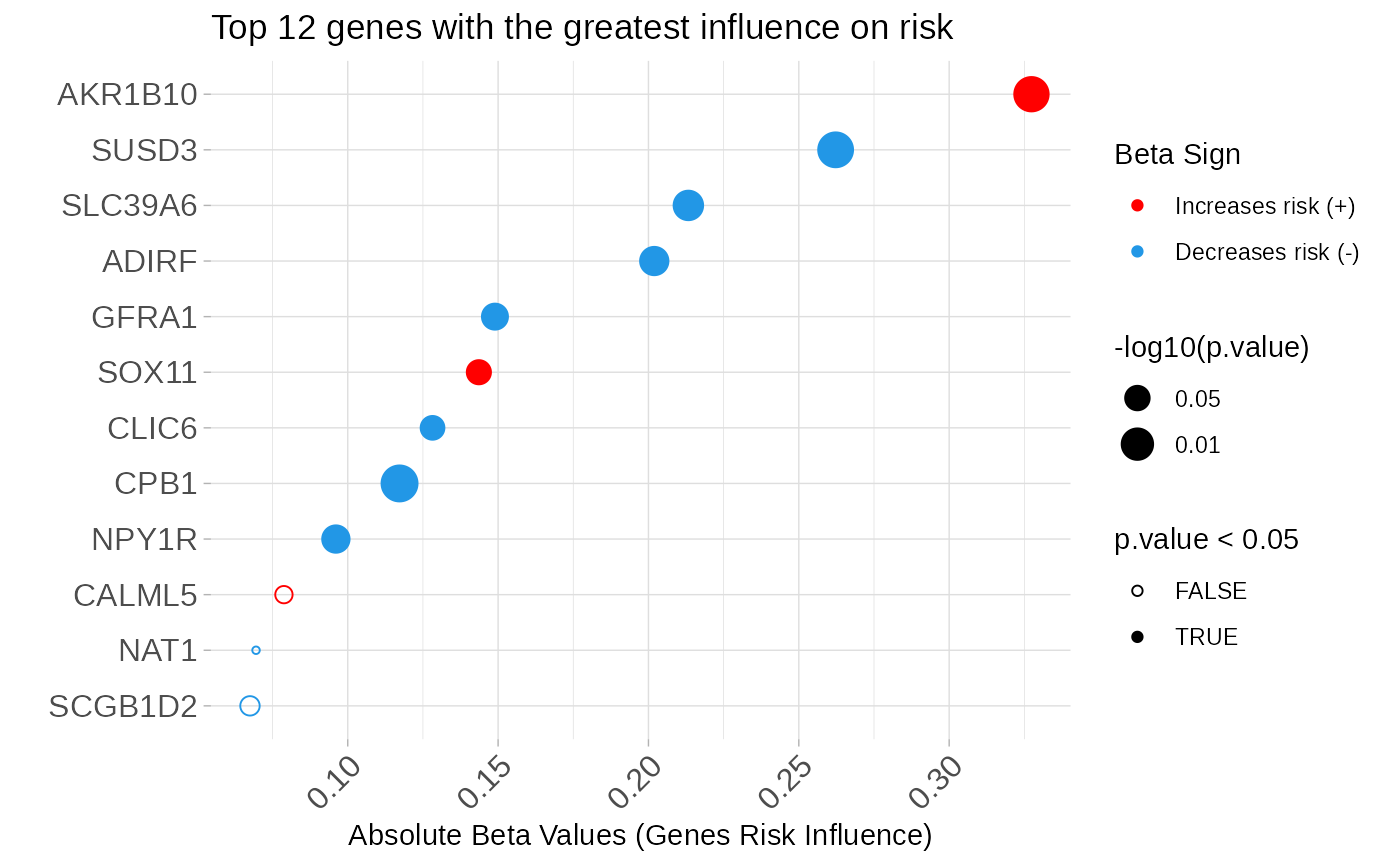
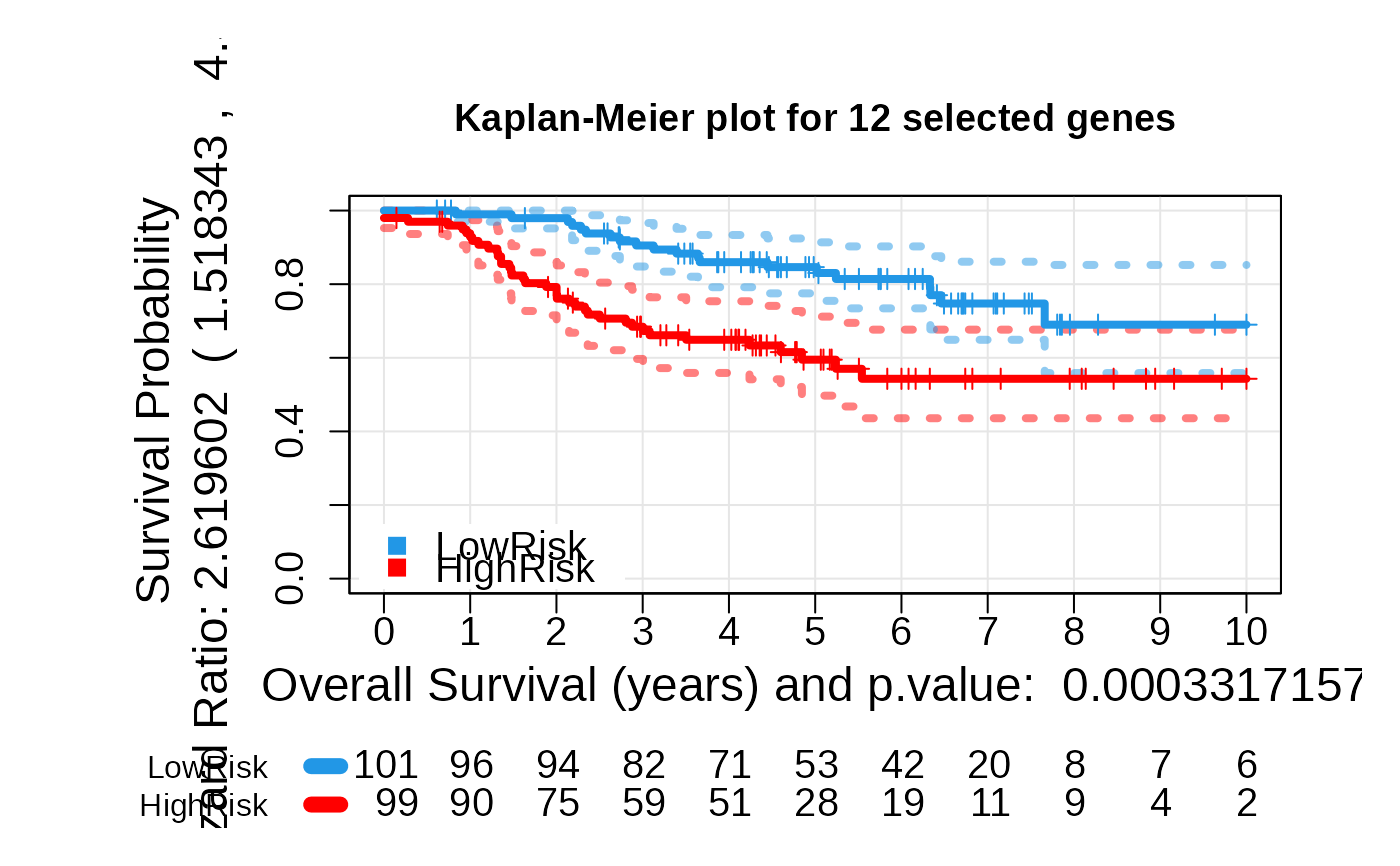
multivariate_risk_predictor <- patientRisk(seBRCA, geneList, time, status,
method = "class.probs")
#> Nested ten fold cross validation: Predicting the risk for each lambda...
#> Nested Cross Validation: optimizing lambda...
#> Risk predicted!
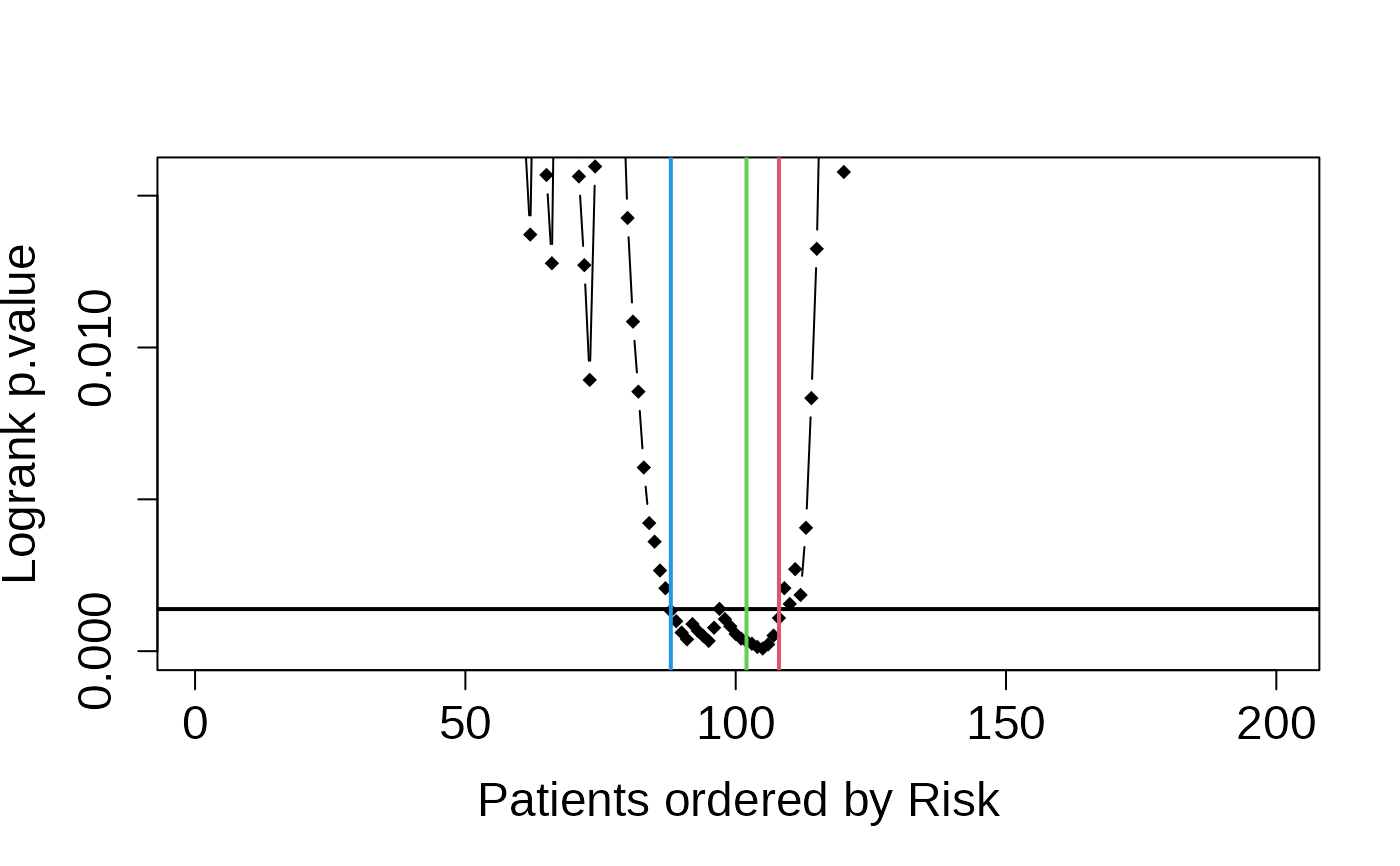
 # Simulate expression data
num_samples <- 20
set.seed(5)
mExprs_testData <- matrix(rnorm(length(geneList) * num_samples,
mean = 10, sd = 3),
nrow = length(geneList), ncol = num_samples)
# Assign row names (genes) and column names (samples)
rownames(mExprs_testData) <- geneList
colnames(mExprs_testData) <- paste0("Sample", 1:num_samples)
set.seed(5)
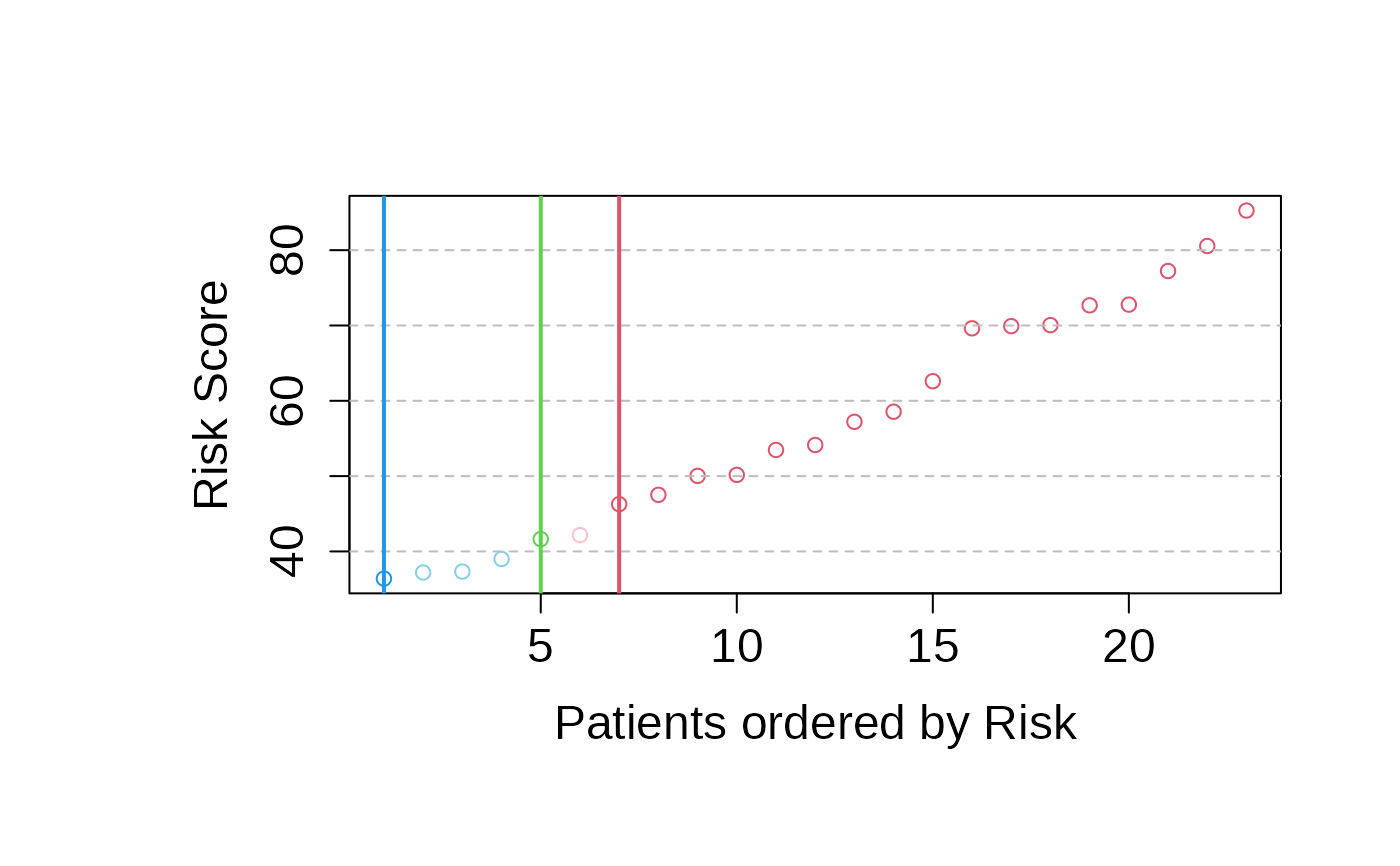
risk_prediction_validation_set <- predict_PatientRisk(
multivariate_risk_predictor,
mExprs_testData)
# Simulate expression data
num_samples <- 20
set.seed(5)
mExprs_testData <- matrix(rnorm(length(geneList) * num_samples,
mean = 10, sd = 3),
nrow = length(geneList), ncol = num_samples)
# Assign row names (genes) and column names (samples)
rownames(mExprs_testData) <- geneList
colnames(mExprs_testData) <- paste0("Sample", 1:num_samples)
set.seed(5)
risk_prediction_validation_set <- predict_PatientRisk(
multivariate_risk_predictor,
mExprs_testData)
 # Example for single patient prediction: Patient fourth is selected.
mExprs_testSingleData <- data.frame(mExprs_testData[, 4])
colnames(mExprs_testSingleData) <- colnames(mExprs_testData)[4]
# Risk prediction for the optimal subset of genes selected by patientRisk()
set.seed(5)
risk_prediction_one_patient <- predict_PatientRisk(
multivariate_risk_predictor,
mExprs_testSingleData)
#> Normalized patient Risk (0 100): 50.0280115373784
#> The patient is categorized as High Risk
#> High Risk interval (46.2794277806291100)
# Example for single patient prediction: Patient fourth is selected.
mExprs_testSingleData <- data.frame(mExprs_testData[, 4])
colnames(mExprs_testSingleData) <- colnames(mExprs_testData)[4]
# Risk prediction for the optimal subset of genes selected by patientRisk()
set.seed(5)
risk_prediction_one_patient <- predict_PatientRisk(
multivariate_risk_predictor,
mExprs_testSingleData)
#> Normalized patient Risk (0 100): 50.0280115373784
#> The patient is categorized as High Risk
#> High Risk interval (46.2794277806291100)